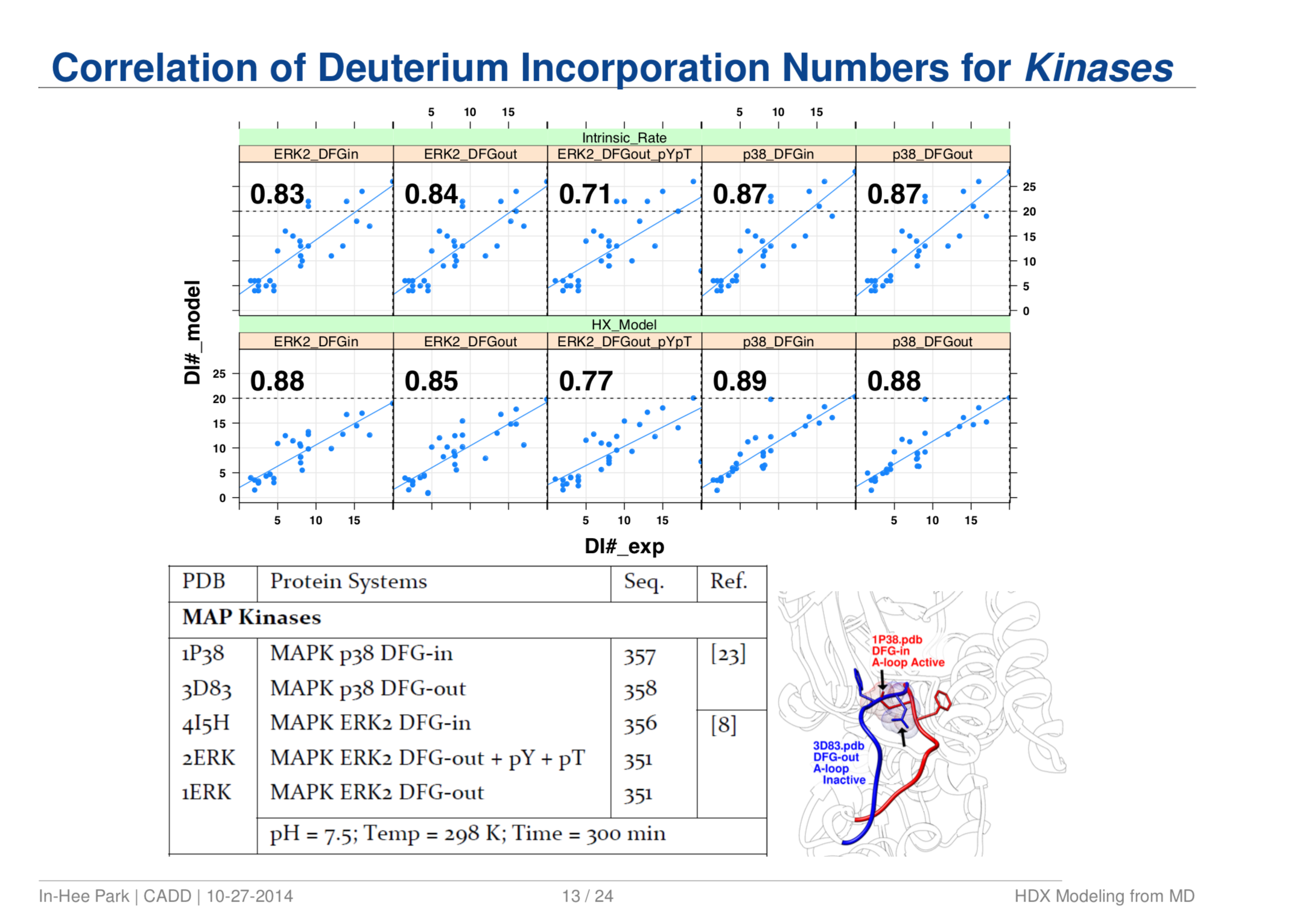
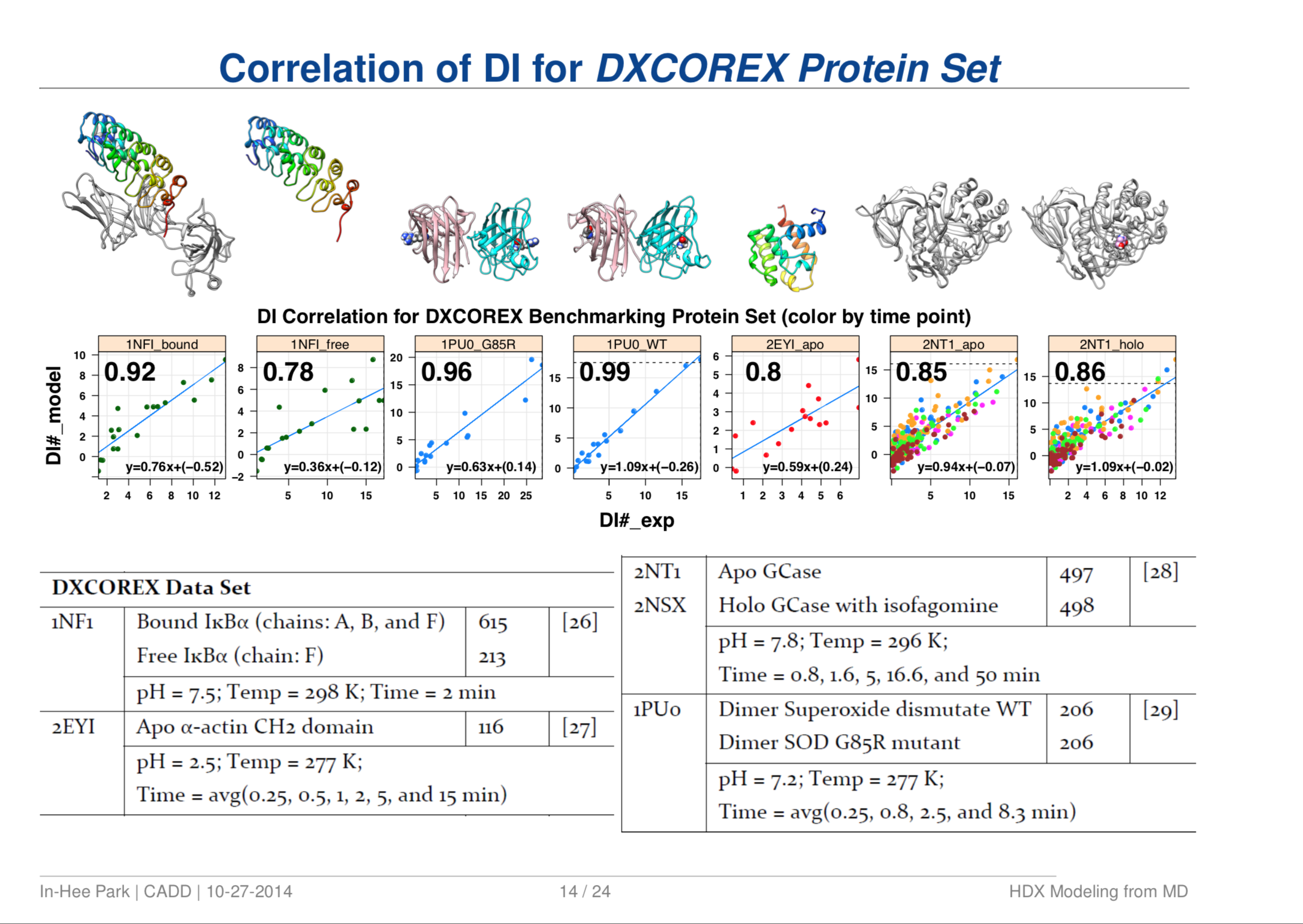
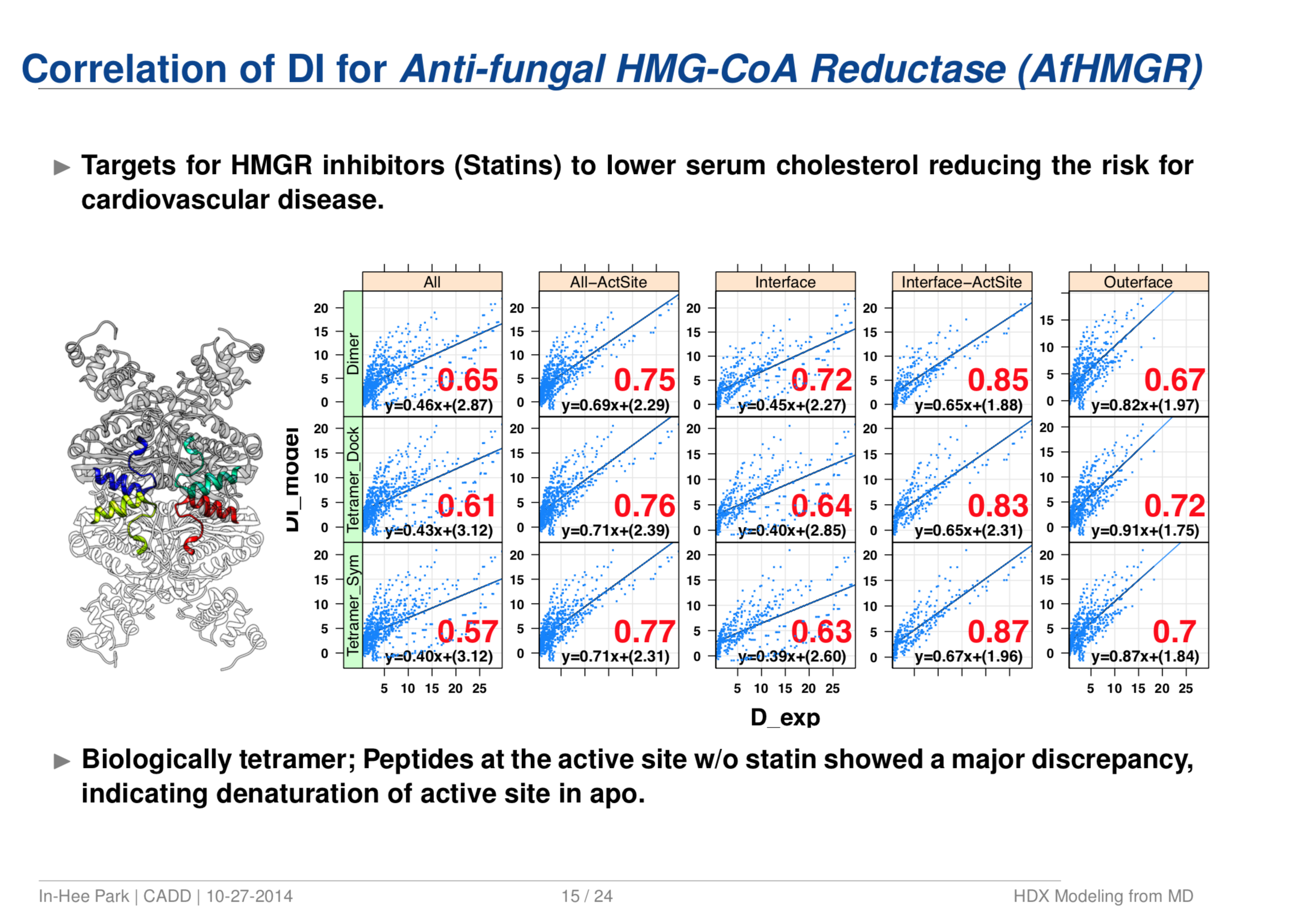
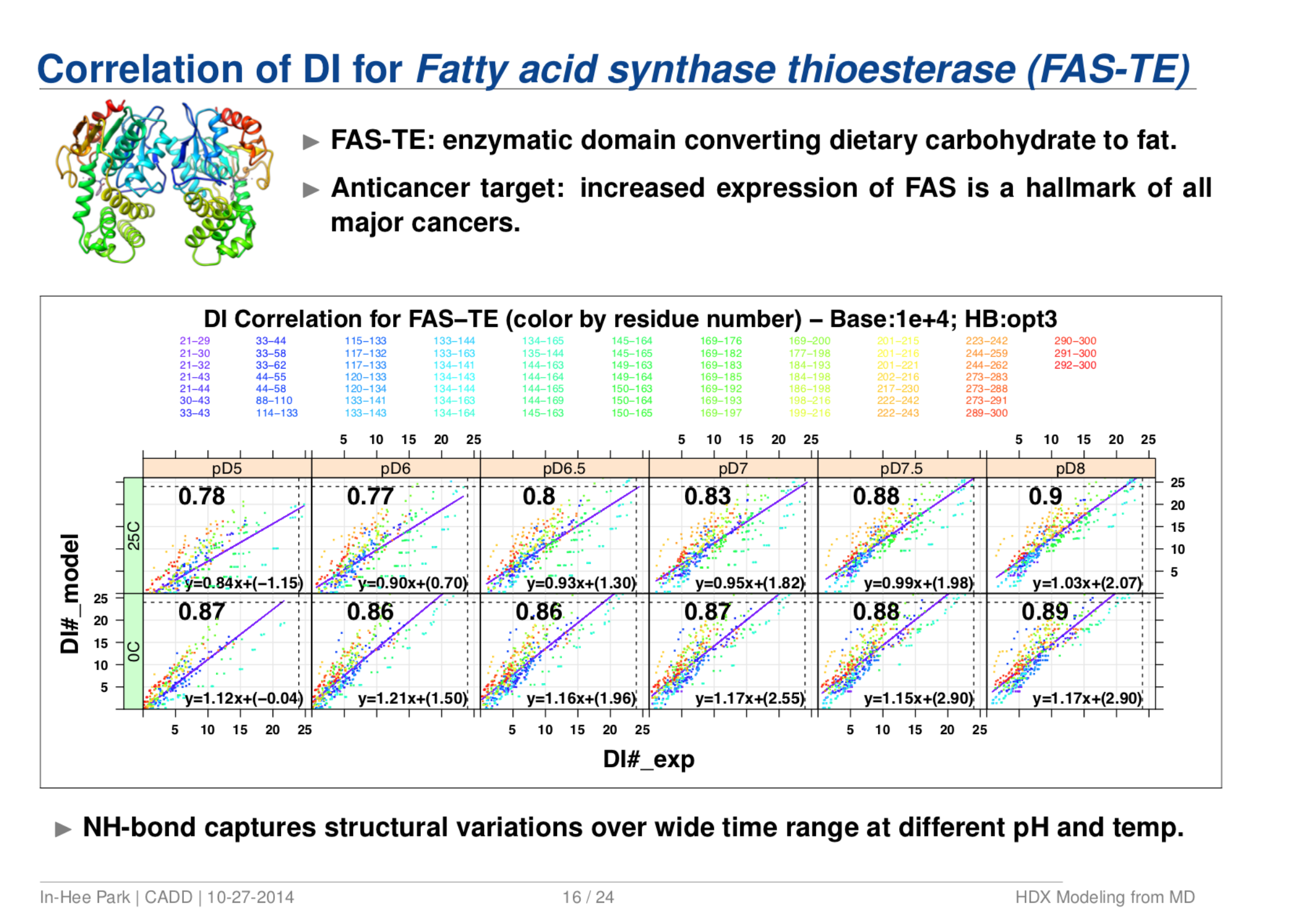
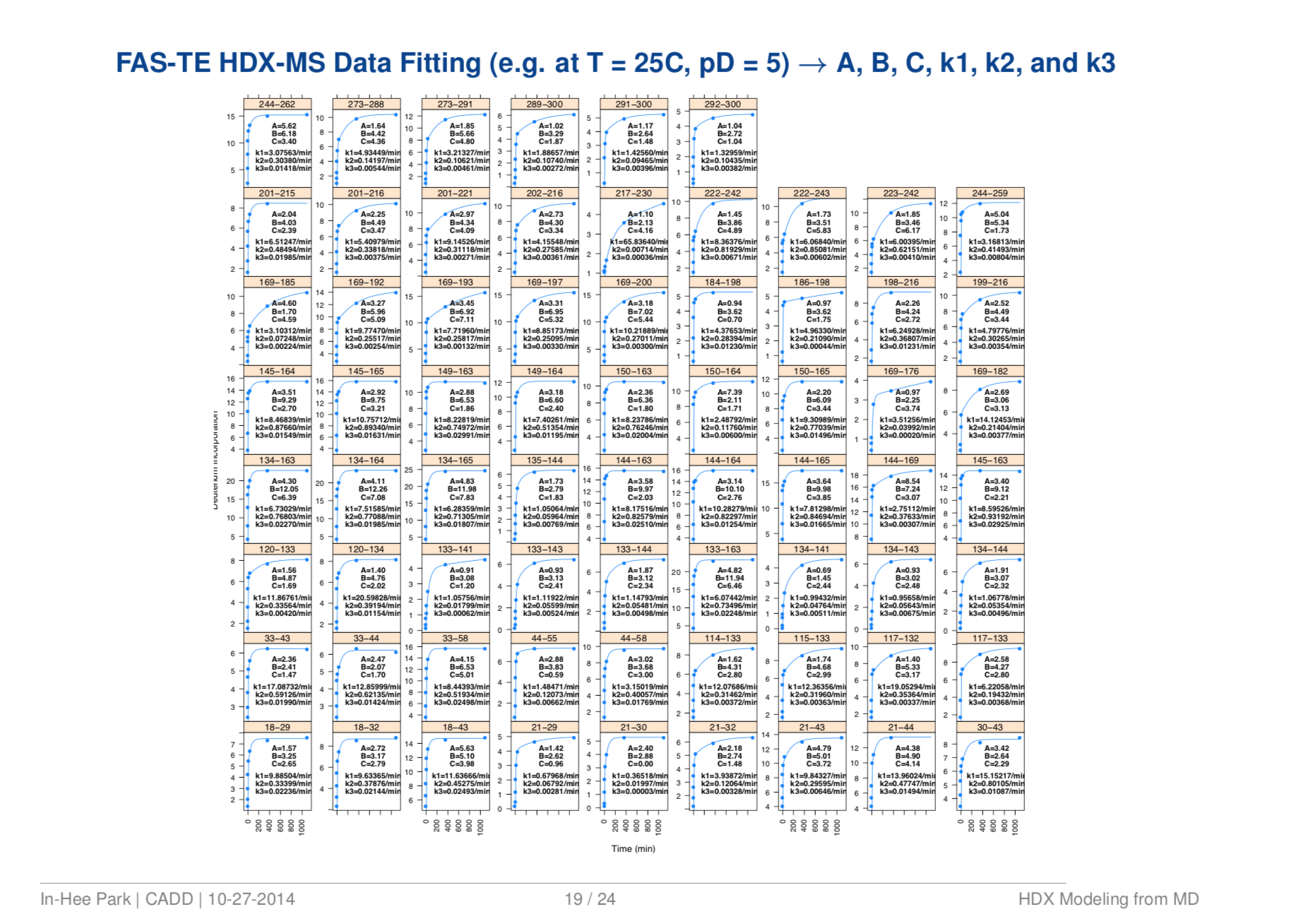
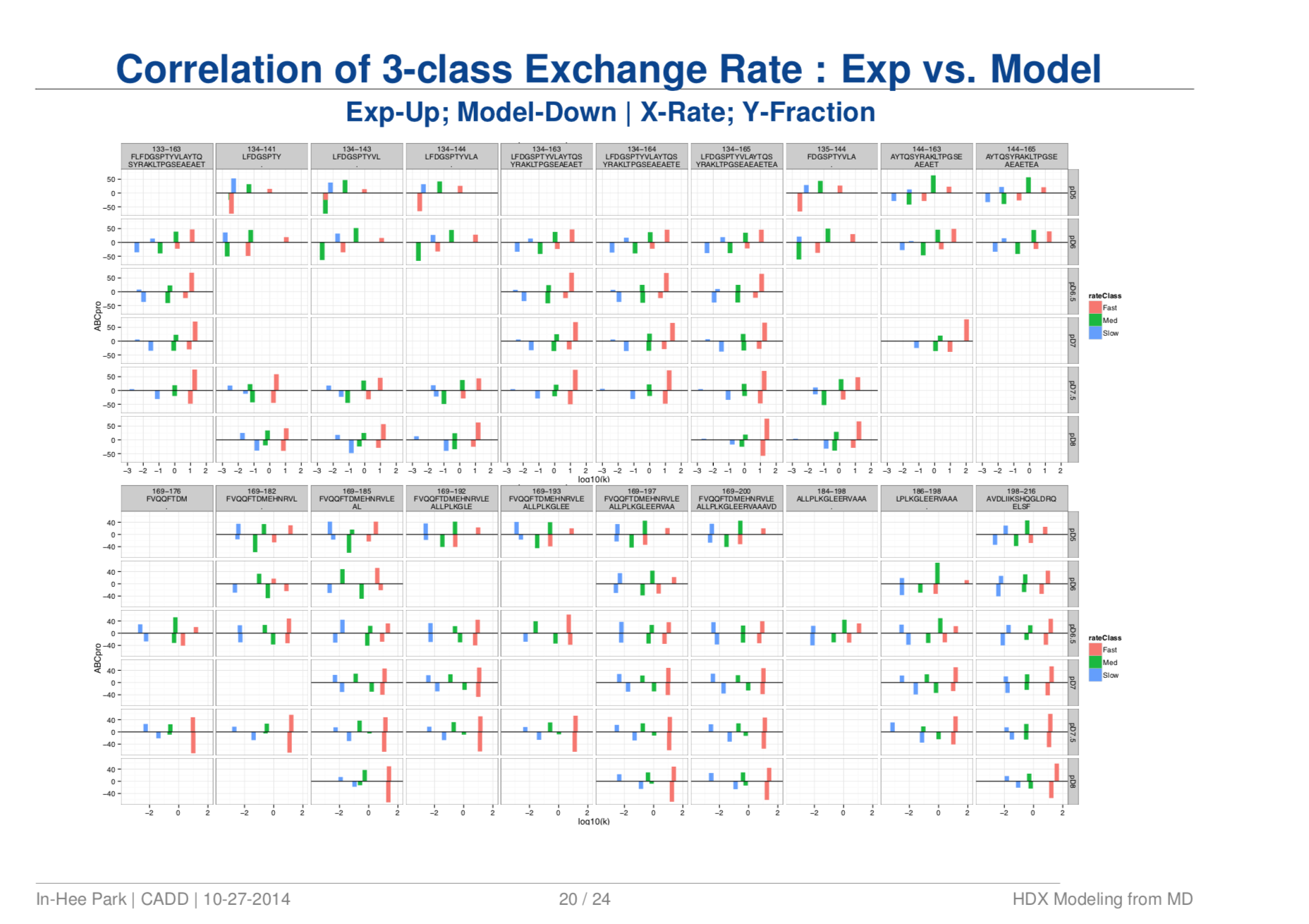
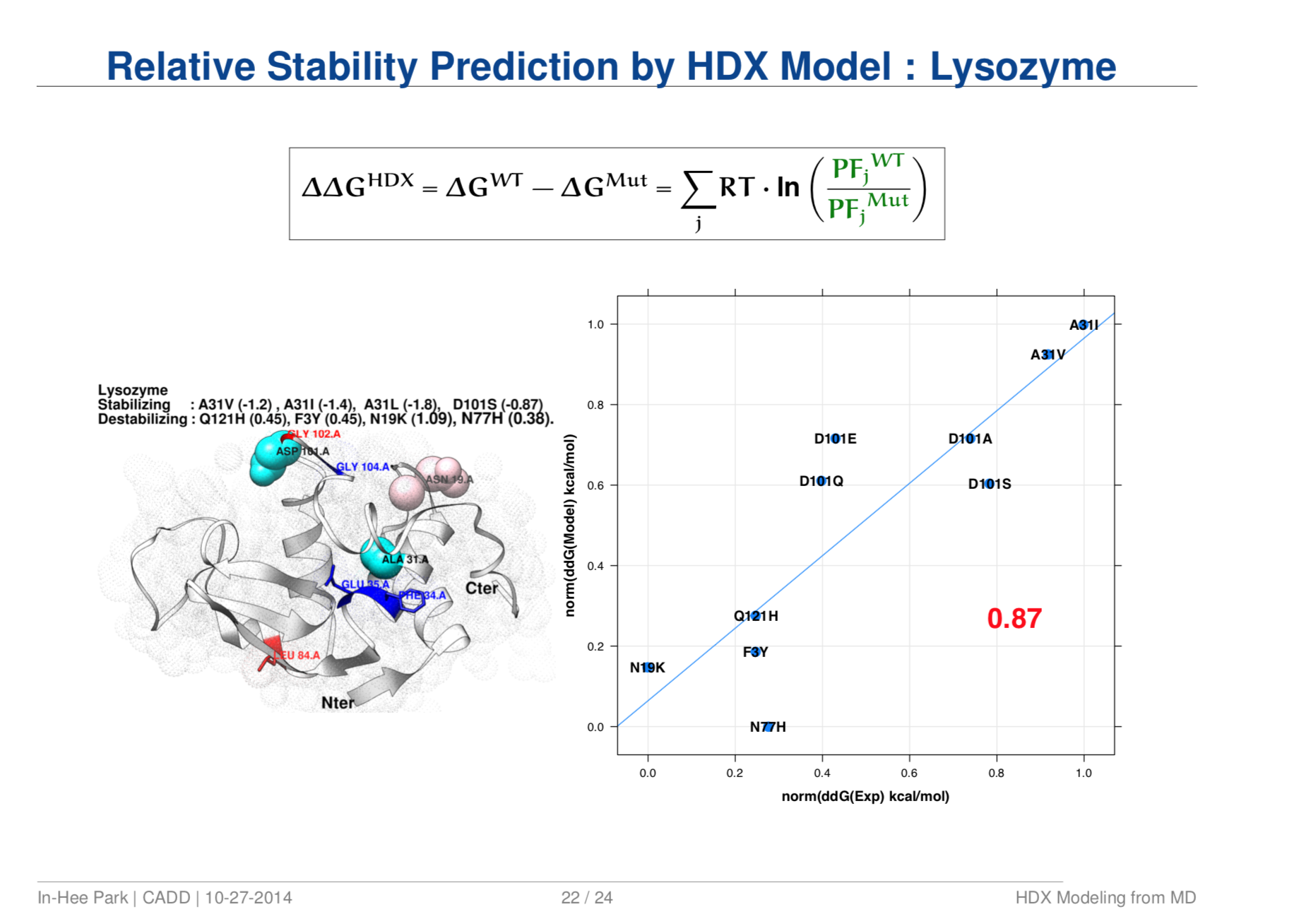
Quantitative Structure Prediction Model for Mass Spectrometry of Biologics from MD Simulation Data
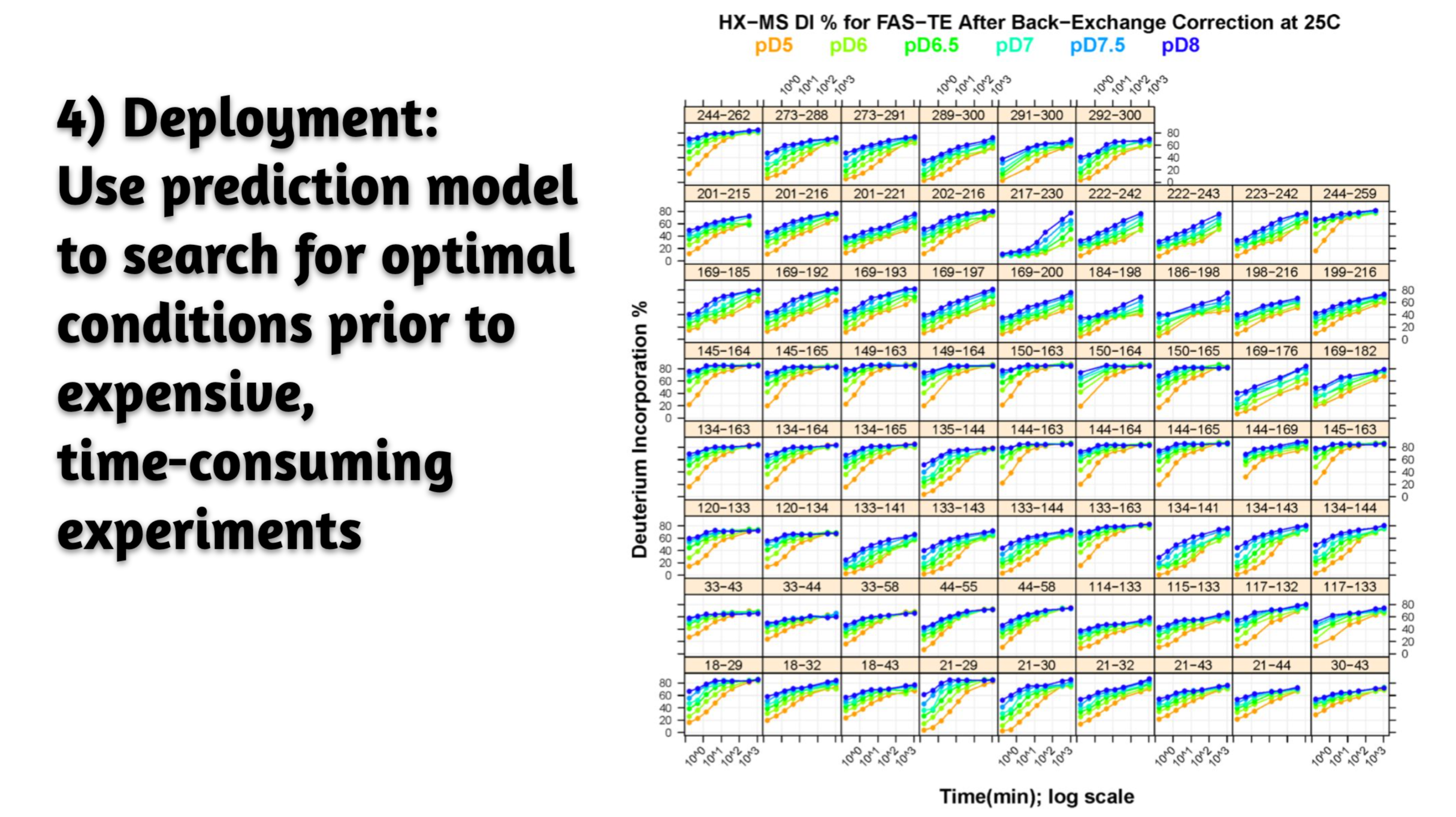
- Problem. Bioanalytics group constantly generated big data for biologics; looking for computational model prediction prior to resource-consuming/labor-intensive mass spectrometry experiment.
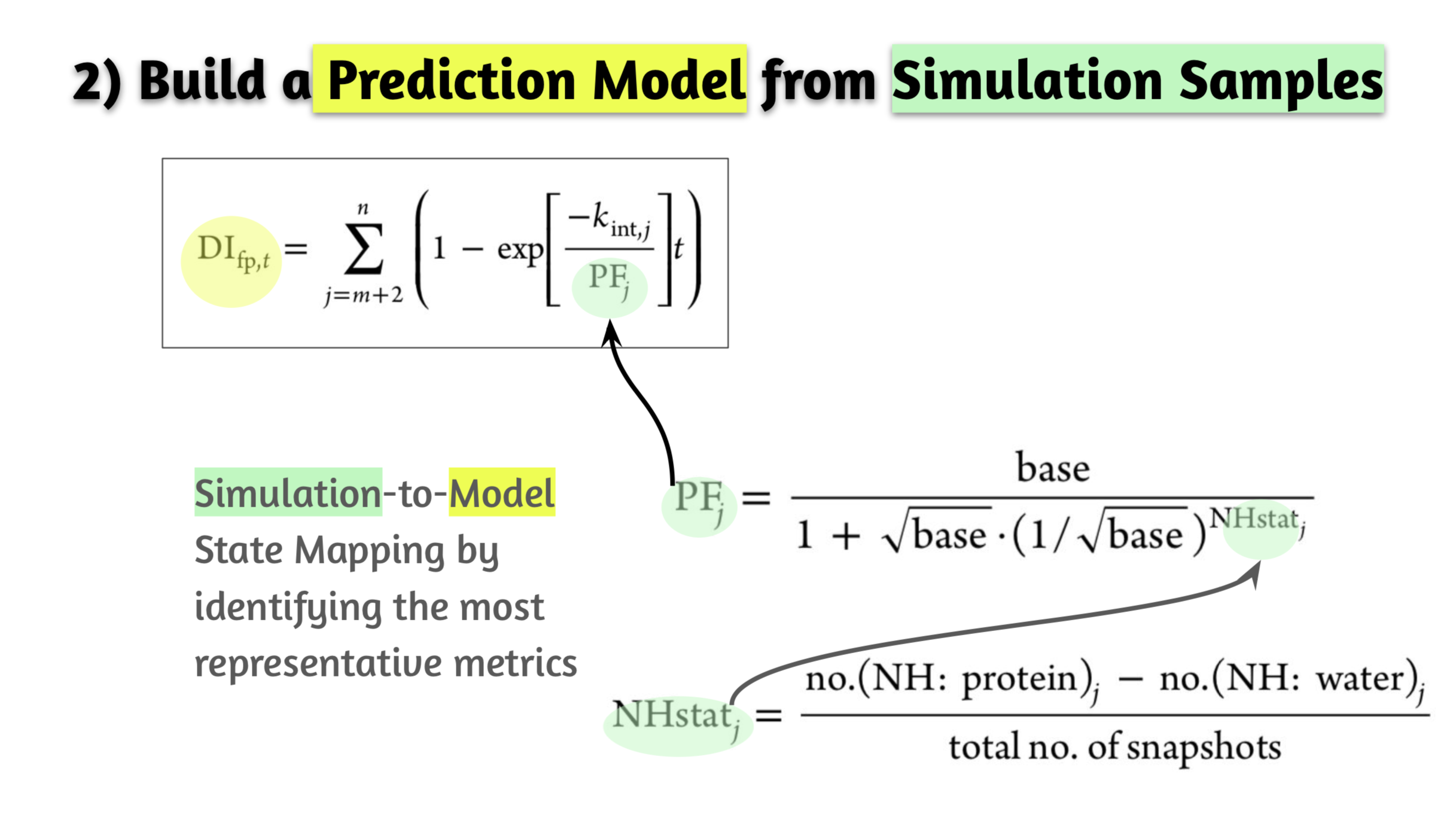
- Action. Initially fully implemented Monte Carlo based COREX algorithm from scratch by referring to scatterd publications & patent documents. Based on the insight gained from the implemented algorithm, developed totally alternative method by accelerating computation time (GPU-enabled MD) and incorporated statistical analysis: dynamic metric idenficiation, growth logistic model construction, simulation-based ensemble data generation, statistical analysis of metric enabling prediction of dynamics at various experimental conditions.
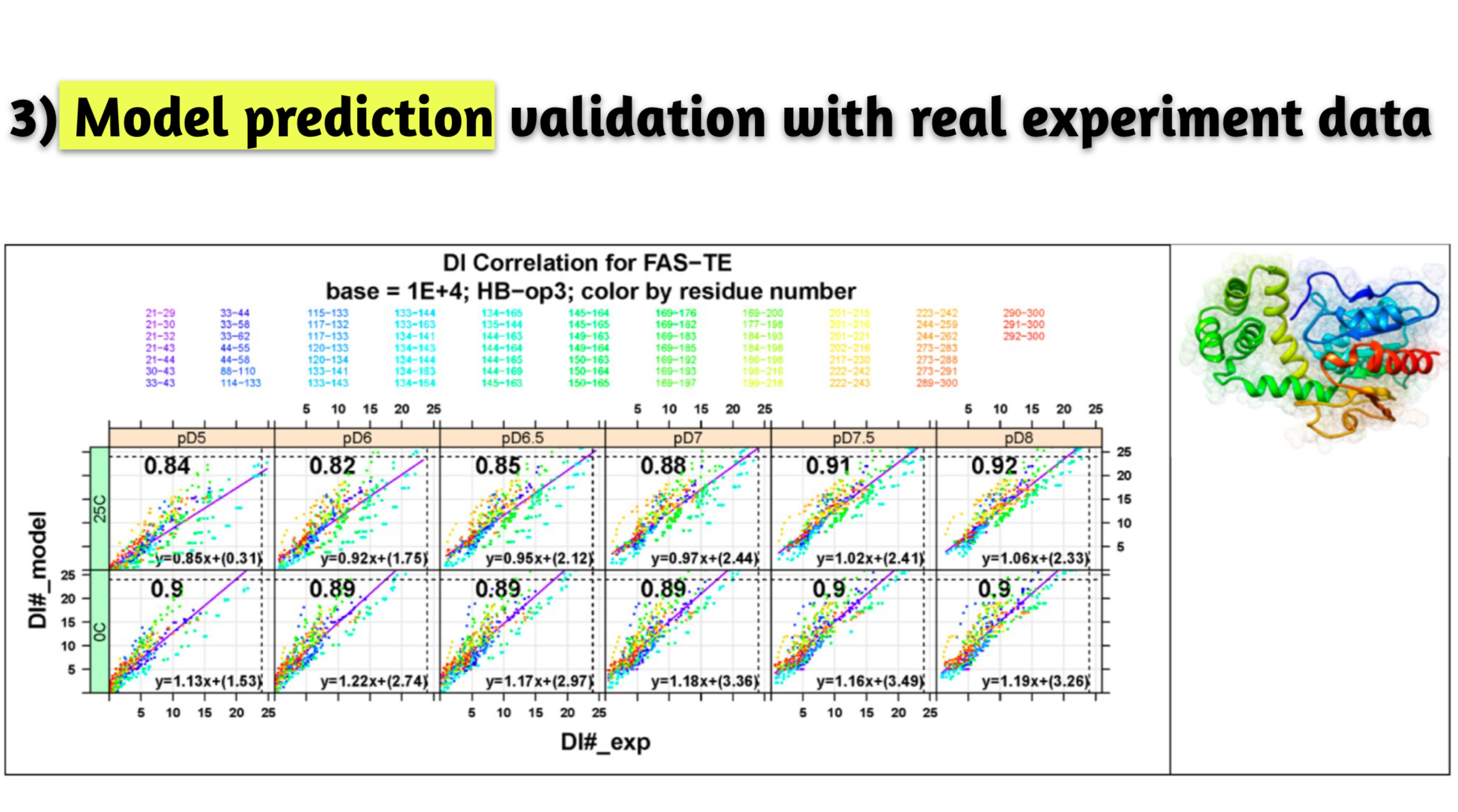
- Result. Turned out to be simple, generic, and robust prediction model with successful benchmark against wide range of biologics systems; aided experimental data interpretation; extended applications to other areas such as stability prediction per mutation.
- Computing. Python, R, SQLite, GPU-enabled MD suite compile/simulation
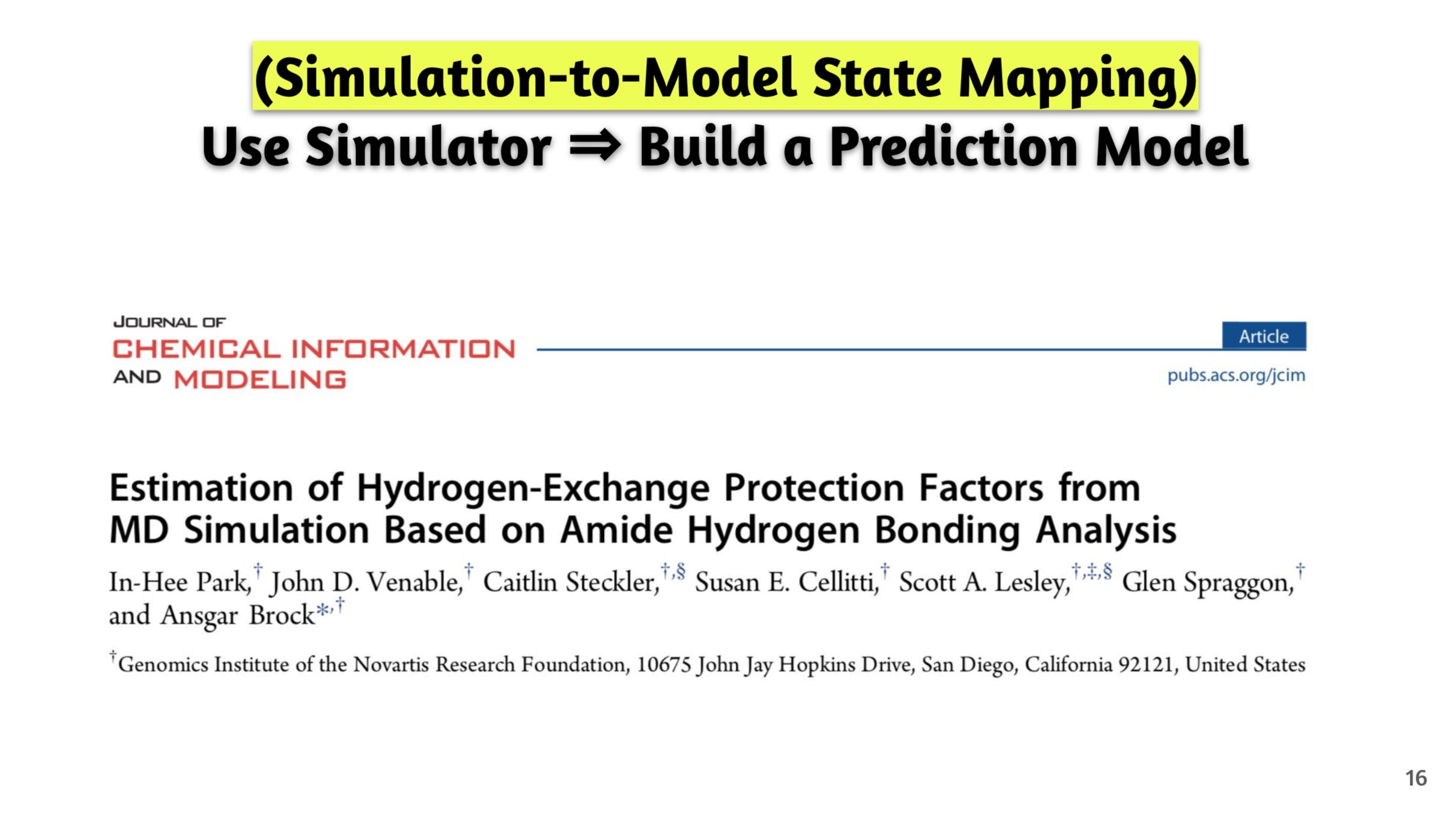
- Publication. Park, I.-H. et al.
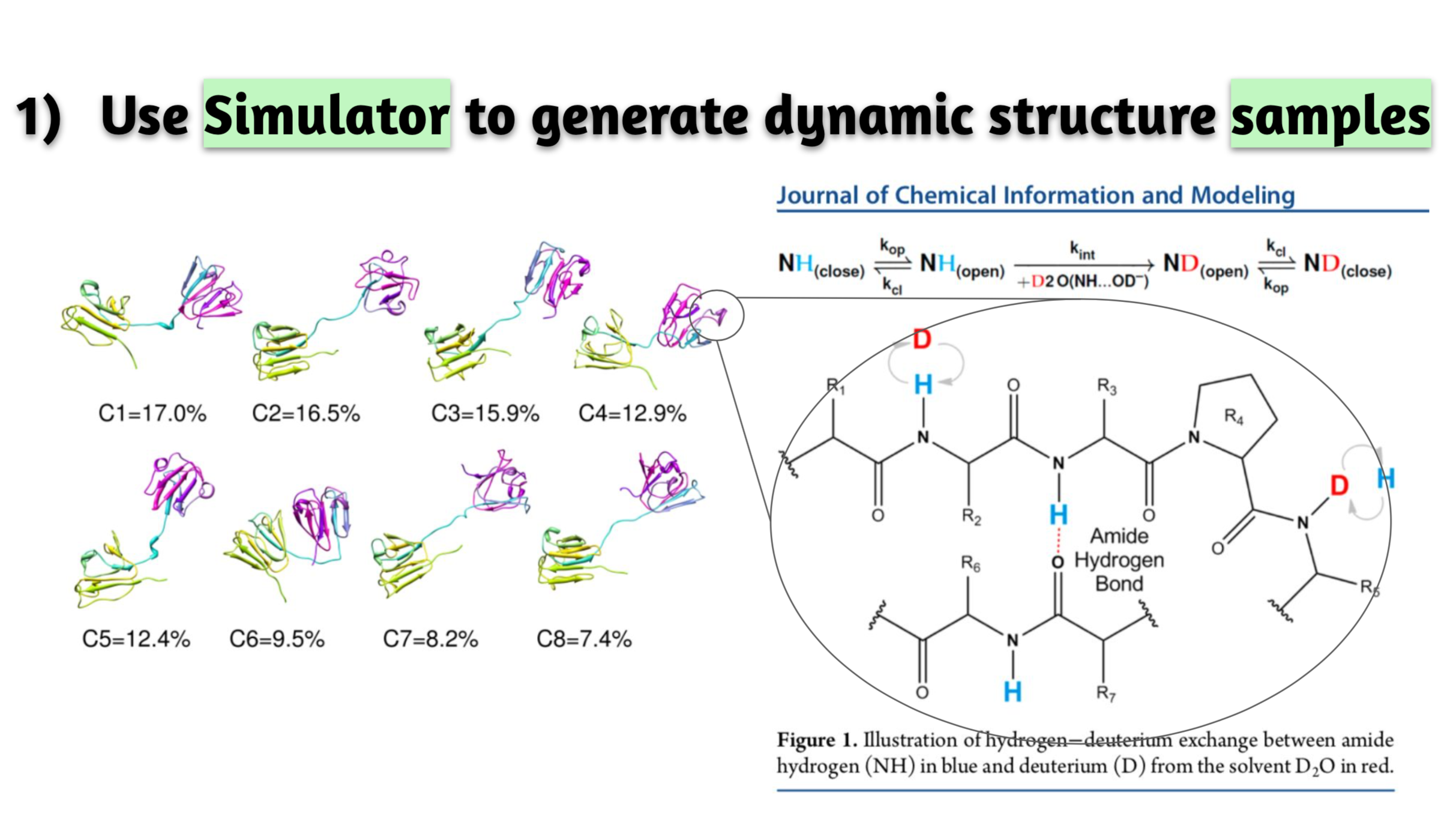
Estimation of Hydrogen Exchange Protection Factors from MD Simulation Based on Amide Hydrogen Bonding Analysis.
J Chem Inf Model. 55(9):1914-25. 2015
High-level Summary
Paper Summary